Speeding up python code with cython pure
Advantages of hybrid languages for numerical computing

Table of Contents
In coding there is a trade-off between writing abstract code, and high performant code. This trade-off is often embodied in different languages. For example, python is considered to be an abstract language whereas C++/C is considered a low level language. Abstract languages allows for rapid prototyping while hiding away the implementation details in packages or modules. A key strategy for abstract languages is providing high level interfaces for low-level implementations allowing for a “best of both worlds” scenario, improving the performance of the abstract language. However, there are cases in which the code written is within the ecosystem of the abstract language, yet the performance needs to increase. For python there are ways to improve the performance of python code. For example, one could use Numba to utilize the Just-In-Time-Compiler (JIT) or use a specialized compiled version of python such as pyston. Cython offers yet another way by compiling special “pyx” files into a shared object that can then be interfaced with from python, creating a hybrid language between C/C++ and python (see my prior post on cython templates). Cython has the advantage of leveraging the existing python stack, and enhancing it with faster run time. It works specifically well if the original python code contains a lot of for loops that cannot be optimized away by other means. Although the cython ecosystem has matured quite a lot in recent years, it requires knowledge from other languages to implement algorithms efficiently. This creates a gap for python developers wanting to have a bit more performance.
A more recent development is compiling existing python code
in “pure” mode in cython. In pure mode, the original code is
left untouched (i.e. the .py file) and the performance
enhancing code is written in a special header file (.pxd).
This has the advantage for increasing the performance of
existing python code while retaining the compatibility with
non-cython users. I recently implemented the forceatlas2
layout in cython’s pure mode to gain a 400 percent increase
over the traditional python code.
In this post, I wish to introduce the reader to cython’s pure mode and test some simple benchmarks to see what kind of performance boosts we can have.
What are header files?
In traditional compiled languages such as C/C++ the source code is split into the implementation and header files. Header files can be thought of as the index of a book, whereas the source files can be thought of the actual content of a book’s chapter. The header file instructs the compiler (and the programmer) on what is present in the implementation and how external code can interface with it. In python, the concept of source and header file does not exist. The closest thing I can think of is showing what kind of class properties a class has. Something similar to,
class Chicken:
mass = 0
height = 0
def __init__(self, name):
self.name = name
Karl = Chicken("Karl")
print(f"My name is {Karl.name} and I weight {Karl.mass} kg")
My name is Karl and I weight 0 kg
Here, the variables mass and height are defined before
the class is inititalized. It tells the programmer that the
class Chicken has two class properties mass and
height. A python programmer would put these variables in
the class constructor as these are individual traits. The
use of init function is general is not the same as this use
case. For the init function may transform the values passed
to the object, whereas here mass and height are clear
class traits.
Cython pure mode
Cython’s pure mode takes an existing python file and
augments it by an header file. Often this header file has
the same name as the original python file but with the
extension .pxd (or definition file). Let’s take an example
function. Assume we want to implement a matrix dot product.
A naive implementation will look something like this:
#file: dot.py
import numpy as np
def dot(A: np.ndarray, B: np.ndarray) -> np.ndarray:
m, n = A.shape
k = B.shape[1]
assert n == B.shape[0], "matrices A and B are not alligned"
output = np.zeros((m, k))
for idx in range(m):
for jdx in range(n):
for kdx in range(k):
output[idx, kdx] += A[idx, jdx] * B[jdx, kdx]
return output
Testing the code for speed we can confirm that this dot product is quite slow.
import numpy as np
A = np.eye(10)
B = np.eye(10)
%timeit -n10 -r10 dot(A, B)
595 µs ± 301 µs per loop (mean ± std. dev. of 10 runs, 10 loops each)
In comparison the numpy implementation is about 200 times faster
import numpy as np
%timeit -n10 -r10 np.dot(A, B)
The slowest run took 26.21 times longer than the fastest. This could mean that an intermediate result is being cached.
7.48 µs ± 16 µs per loop (mean ± std. dev. of 10 runs, 10 loops each)
Let’s say we really like our dot implementation, but we wish the python code was a bit faster. For cython pure mode, we would need to augment the py file with pxd file. The header file would need to contain
- The functions name
- The functions return type
- The functions input arguments
We create the file dot.pxd and fill in the header as follows:
# file: dot.pxd
# distutils: language = c++
# or c if you want c
import cython
@cython.locals(m = size_t, n = size_t, k = size_t,
idx = size_t, jdx = size_t, kdx = size_t,
output = double[:, ::1])
cpdef double[:, ::1] dot(double[:, ::1] A, double[:, ::1] B)
What exactly happened here? The `cython.locals` tells the
compiled which variables are declared inside the dot
function in the py file. It tells the compile what the types
are. The function description is exposed through cython with
the cpdef statement. This exposes the function to calls
from python in the .so file. Next the return type is
given. Here a memoryview is used. These are raw buffers that
can be directly converted from numpy. They have faster
access than normal the normal numpy arrays. Using memory
views has the disadvantages that the numpy methods that are
defined on the array are disable. For example one cannot
call A.sum or A.mean or A.reshape anymore when using
memory views. An alternative would be to use `np.ndarray`
however these are still considered to be python objects and
therefore do not have increased speed compared to the
memoryviews.
We then compile the source code either from a setup file(not covered but preferred in modules) or directly from the commandline with
cythonize build_ext --inplace dot.py
which produces an .so file. Let’s test the speed!
from dot import dot as cydot
%timeit -n10 -r10 cydot(A, B)
4.11 µs ± 557 ns per loop (mean ± std. dev. of 10 runs, 10 loops each)
Which is a nearly 200 percent increase in speed! Importantly, it is nearly on par with the numpy’s implementation!
Benchmarks
Let’s benchmark the results to see how the different implementations scale with matrix size.
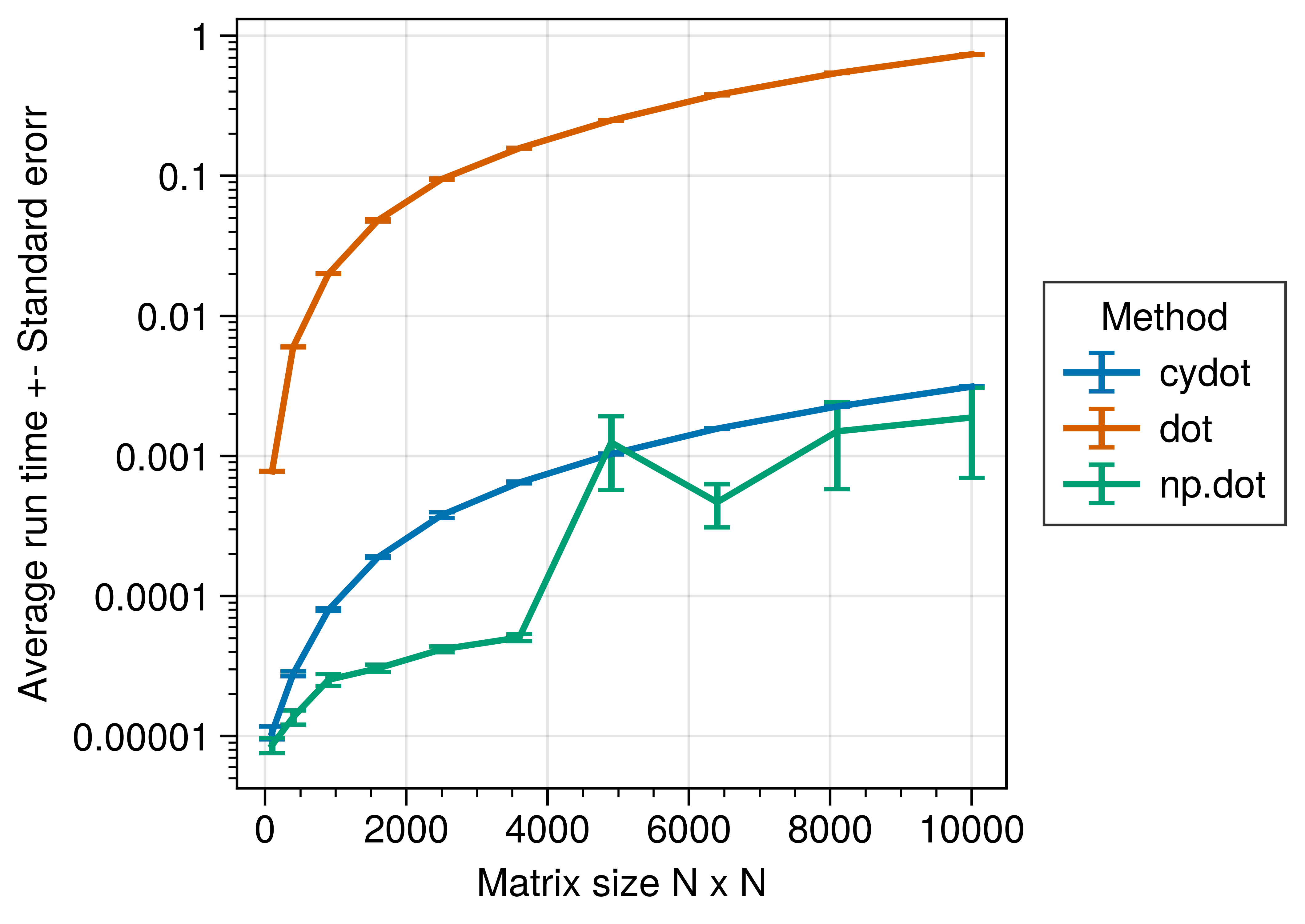
Summary
Cython can be used to speed up existing python code. The preferred way is to use cython pyx files with pxd header files. An alternative is to use cython pure mode to speed up existing python code. Pure mode allows for smoother collaboration with numerical experts and python developers.